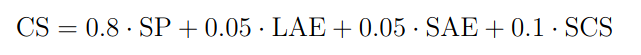Task Description¶
The aim of the challenge is to develop Universal Lesion Segmentation (ULS) models for thoracic-adbominal CT scans. These models should be able to segment and measure the various types of lesions Radiologists see during clinical practice. For the proposed pipeline, first a lesion is identified by a Radiologist and clicked on, then a volume-of-interest (VOI) can be cropped around this location and the lesion is segmented. Lesion characteristics such as the long- and short-axis measurements can then be extracted for further evaluation using e.g. RECIST.

During training and evaluation participants will be provided VOI's of size 256x256x128, in the original scan spacing. This size allows for the largest lesions to be encapsulated completely in the VOI. Where necessary the data has been padded (using the minimum array value -1) to construct the volume. Although the data is in this fixed format we also provide code to extract lesions and build the training data from scratch with different parameters. Participants are also free to crop smaller volumes out of this data or resample it during preprocessing, before evaluation on the original volume size and spacing.
In our discussions with radiologists we have found that for a ULS model to be clinically relevant it needs to be robust, precise and fast.
In order to determine robustness we have curated a test set containing various lesion types, sizes, areas imaged, slice thicknesses, spacings and contrast uses. All our test lesions were originally measured during oncological workup by a radiologist, which we take as a measure of relevancy of these particular lesions. We also measure the segmentation consistency of models by sampling multiple lesion click locations based on which inference volumes are cropped, as discussed in the metrics section of this page.
We evaluate segmentation performance and the bi-dimensional measurement error of the developed models. Currently in clinical practice, often only uni- or bi-dimensional measurements are used to track lesions. However, since it is easier to optimize for these simple metrics without taking into account the actual shape of the lesion, and since accurate lesion volume predictions can be used to provide additional clinical markers in the future, we believe including segmentation performance metrics gives more insight into model performance.
Inference speed is also a serious consideration for being able to use a ULS model in the clinic. If a radiologist needs to wait multiple seconds for a lesion they clicked on to be segmented it slows down the workflow considerably. As detailed in the resources section below, participants will have limited time per volume to segment a lesion. Compared to other segmentation challenges this means participants will have to carefully consider model size and complexity, and ensembling multiple large models is discouraged.
Evaluation¶
There are two tracks for model evaluation, one for challenge participants and the other for non-participants. Non-participants can benchmark their models on our test set, but are not eligible to win the challenge. We have additional requirements for challenge participants as detailed in the rules.
The ULS23 test set will not be made public. In order to evaluate your model you will need to submit a docker container containing your pre- and post-processing code and your network. This container will be run as an algorithm on GrandChallenge. We will provide documentation on how to construct such a container and will provide a base image which contains the necessary code to prepare the correct inputs and outputs.
Resources¶
For inference a T4 GPU with 16GB of VRAM, 8 CPU and 32GB of CPU RAM is available (the g4dn.2xlarge instance on AWS). Each job will have a maximum runtime of 9 minutes. During this time preprocessing will need to be run, the model weights need to be loaded, inference concluded and finally postprocessing and exporting needs to happen. Each job will need to run inference on 100 VOI's of 256x256x128 pixels in this timeframe. A lesion foreground pixel is always centered in the volume. Participants can crop or resample volumes, but will need to return output segmentations in the same format as the input images.
Metrics¶
Segmentation performance (SP) will be calculated by taking the mean of the Sørensen–Dice coefficient over the test set.
Since it is often used as a metric in the clinic, we also evaluate the long- and short-axis errors (LAE and SAE). For this the long- and short-axis diameters of the lesion at its greatest axial extent will be determined from the 3D segmentation mask. We take the short-axis as the maximum lesion diameter perpendicular to the long-axis. These measurements are then compared to those extracted from the ground truth. We will use the symmetric mean absolute percentage error (SMAPE) for this evaluation. This metric ranges from 0 to 1, where a score of 0 is achieved when both measurement are the same size. If a model doesn't provide a prediction for a case this leads to the maximum error of 1 being used for that case.

Furthermore, a subset of test lesions will be included multiple times using different lesion click locations. This results in slightly different scan context for each cropped VOI. We check whether the model outputs similar predictions using these different click locations by comparing the Sørensen–Dice coefficient of the re-aligned segmentation masks. This is taken as the segmentation consistency score (SCS). A model robust to click location variation should have highly similar output segmentations.
For the final ranking, metrics will be aggregated for the Challenge Score (CS) as: